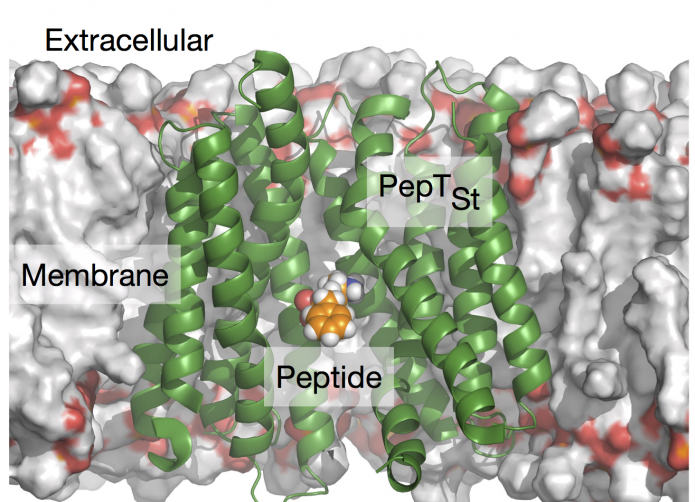
PepT1 is a nutrient transporter found in the cells that line your small intestine. It is not only responsible for the uptake of di- and tai-peptides, and therefore much of your dietary proteins, but also the uptake of most β-lactam antibiotics. This serendipity ensures that we can take (many of) these important drugs orally.
Our ultimate goal is to develop the capability to predict modifications to drug scaffolds that will improve or enable their uptake by PepT1, thereby improving their oral bioavailability.
, just published online in the new journal Cell Chemical Biology (and free to download, thanks to ), we show that it is possible to predict how well a series of di- and tai-peptides bind to a bacterial homologue of PepT1 using a hierarchical approach that combines an end-point free energy method with thermodynamic integration. Since there is no structure of PepT1, we then tried our method on a homology model we have published in 2023. We found that method lost its predictive power. By studying a range of homology models of intermediate quality, we showed that it is highly likely an experimental structure of hPepT1 will be required for in silico accurate predictions of transport.
This is the second paper that Firdaus Samsudin has published as part of his DPhil here in Oxford.